bioView3D volume renderer is an open source and cross-platform application intended for bio-scientists to visualize fluorescence imagery in 3D. It supports images with up to 5 dimensions (3D + time + channels). bioView3D runs on Windows, MacOS X and Linux. It's based on OpenGL and will run smooth on modern hardware. bioView3D supports popular image stack formats like TIFF, Olympus Fluoview TIFF, Olympus OIB, Zeiss LSM, Zeiss ZVI, OME-TIFF, BioRad PIC, etc.
bioView3D has all the necessary tools embedded - image interpolation and channel remapping, it can show up to 7 simultaneous channels. The volume is constructed using meta-data extracted from image files so it's physically accurate and seamless to users. It's also possible to export 3D images, screens and videos into files.
External links:
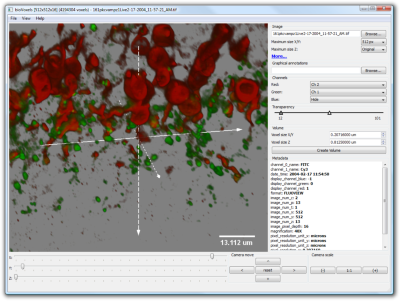
The navigation over the 3D sample is simple and can be done by only using a mouse and it's buttons. Gestures allow full navigation capabilities using a multi-touch track pads.
Windows notes: If you only have a normal 3 button mouse, make sure to enable [View -> Use mouse wheel to zoom], it should be selected by default
MacSOX notes: Make sure that the option [View -> Use mouse wheel to zoom] is disabled, this will allow 2D panning, it should be de-selected by default
There are two ways to export movie: the path based, in which the user can define visualization locations and number of frames and the movie will be interpolated accordingly. The second is the live mode in which frames are stored with user defined frequency. Movie export supports a wide variety of formats and can generate Quicktime, Adobe Flash video, AVI, MPEG4, OGG, Windows Media Video, Multi-page TIFF, etc.
Graphical annotations that the bioView3D can visualize are simple and rather intuitive XML files specified and used by the Bisque database. Additionally, it can read files produced by the Digital Notebook software. These XML files store annotations in hierarchical trees with leaf nodes being graphical primitives like "point" or "line" and other nodes being grouping nodes with humanly meaningful types and names. Types can be something like: "amicron cell" and names something like "cell 1". In order to read more about these annotations refer to the Bisque paper published in Bioinformatics, 2010. The section 2 in the supplementary material describes details and gives examples of such files.
bioView3D does not load multi-file images for the reason that most modern microscopes produce single file 5D images that contain all the necessary image data and metadata. Although it is possible to construct a single 5D image file from multiple files using BioImage Convert utility. It is available for Windows, MacOSX and Linux and supports many image formats. I could suggest to create images in OME-TIFF image format for it is an open and widely supported format, it is a normal multi-page TIFF file containing an XML description of image geometry in the description tag. The following command line would construct a 3D Z stack from 3 input images with resolution of X 0.2um Y 0.2um Z 1.0um T 1.0s:
imgcnv -i in1.tif -i in2.tif -i in3.tif -o out.ome.tif -t ome-tiff -geometry 3,1 -resolution 0.2,0.2,1.0,1.0
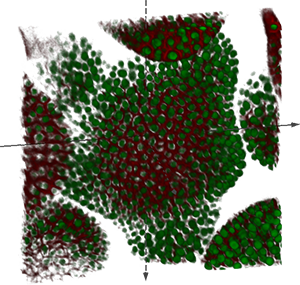
The first image shows fixed rabbit retina cross-section, acquired using an Olympus FV-500 laser scanning confocal microscope and a courtesy of Geoff Lewis and the Fisher's Laboratory at MCDB, UCSB. The sample is stained with GFAP: Cy5 (Labels glial cells), BrdU : Cy3 (Stain incorporated into dividing cells going through S-phase (DNA synthesis) at time of administration), Mib-1: Cy2 (Labels dividing cells in all phases). The physical image dimensions are 127x127x12.6 microns (512x512x51 voxels) and the voxel size is 0.248592x0.248592x1.0 microns. Image format: Olympus Fluoview TIFF.
Browse: [using web browser]
Download: [Original image], [Associated graphical annotations]
This 4D image shows mitosis (cell division) stages Metaphase-Anaphase by fluorescent tagging of mitotic spindles, courtesy of Alphan Altinok, Caltech. The physical image dimensions are 45.9x45.9x12.9 microns (256x256x14 voxels) with 68 time ponits and the voxel size is 0.179297x0.179297x0.928571 microns. Image format: Olympus OIB.
Browse: [using web browser]
Download: [Original image]
Linux and Mac, as described in <http://kb.mozillazine.org/Register_protocol>
Windows
The preferred way is to use the windows installer that does it automatically.
Or use the techinque described in: <http://kb.mozillazine.org/Register_protocol>